Search
Sharing Knowledge improves Knowledge... Knowledge should come at as less cost as possible.
Posts
Showing posts from March, 2017
Posted by
Varun C N
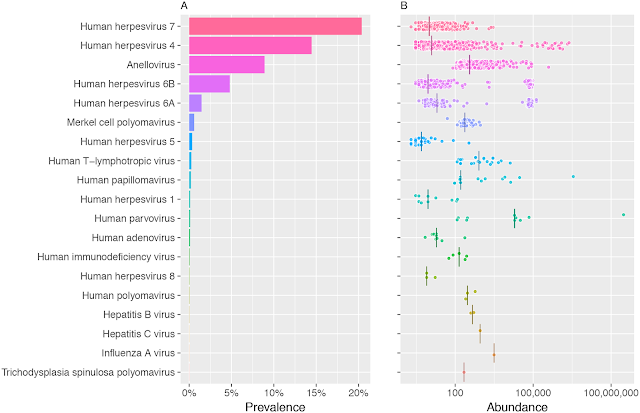
Human Blood Virome
- Get link
- Other Apps
Posted by
Varun C N
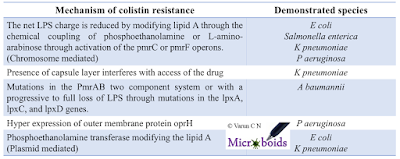
MCR gene variants
- Get link
- Other Apps

Posted by
Varun C N
Bedaquline and Delamanid resistance
- Get link
- Other Apps

Posted by
Varun C N
WHO global PPL for antibiotic development
- Get link
- Other Apps
Posted by
Varun C N
BtB# 12- Why is ID a Requirement in Clinical Microbiology
- Get link
- Other Apps